Stanford Researchers Introduce PEPSI: A New Artificial Intelligence Method to Identify Tumor-Immune Cell Interactions from Tissue Imaging
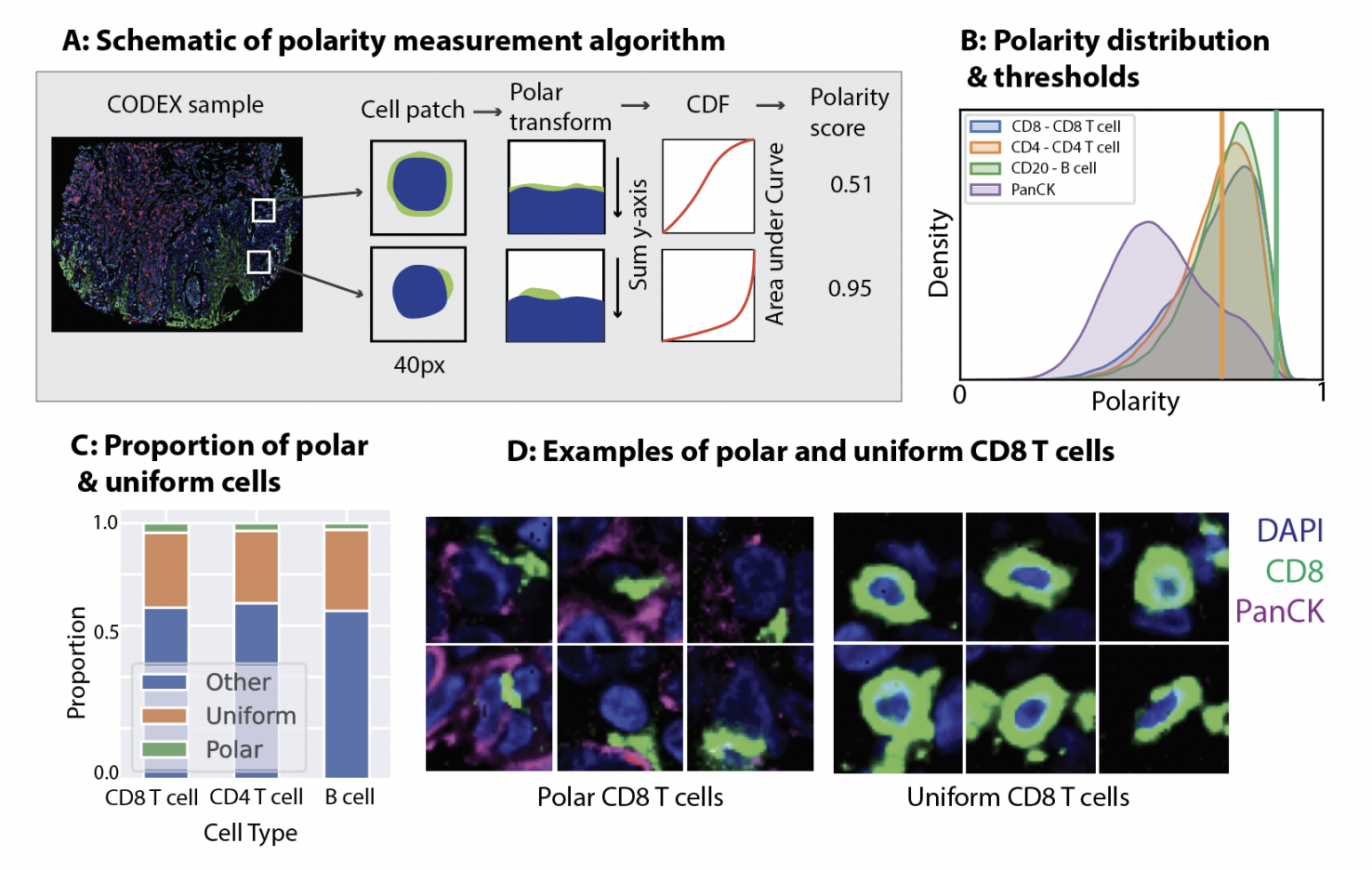
Subcellular protein localization is key to unlocking the intricate details of cellular function, particularly within the complex milieu of tumor microenvironments. While providing a broad view, traditional proteomics methods often miss the nuanced, sub-cellular information crucial for a complete understanding. Researchers have noted that current techniques, such as immunofluorescence, tend to aggregate protein expressions within cells, losing potentially significant morphological signals. This general approach results in an inability to differentiate cells with varied protein localization, a critical factor for understanding immune responses in cancer.
One significant challenge in spatial proteomics is accurately analyzing and quantifying protein localization within cells, especially in tumor microenvironments. The current methodology, primarily centered around immunofluorescence, generally averages out protein expressions across single cells, thereby glossing over sub-cellular details. This not only results in a loss of valuable information but also impedes the identification and differentiation of cells based on their subcellular protein expression patterns. The need for a method that can discern these subtleties is crucial for a deeper understanding of cellular functions due to various stimuli, including immune engagements.
Spatial proteomics techniques like immunofluorescence and immunohistochemistry have been the cornerstone in understanding the spatial structure of tissues at a subcellular level. However, these methods have a notable limitation: they tend to aggregate and average protein expression within single cells, leading to a loss of critical sub-cellular and morphological signals. As a result, the methods fall short in differentiating cells that exhibit differential protein localization, a vital aspect in deciphering cellular functional states, particularly in the context of tumor-immune interactions.
Researchers from Stanford University and Enable Medicine have developed PEPSI (Protein Expression Polarity Subtyping in Immunostains), a novel methodology for measuring subcellular protein localization. This technique is innovative in characterizing the tumor microenvironment by identifying distinct immune cell states. PEPSI operates by computing the polarity of cell surface biomarkers from immunofluorescence imaging data. It classifies cells into subtypes based on their surface protein marker polarity, such as polarized and uniform, offering a refined perspective on cellular behavior.
The methodology of PEPSI involves a detailed, step-by-step process. It begins with the polar transformation of the immunofluorescence signal relative to each cell’s centroid, effectively measuring surface protein polarity. This process allows for identifying cells with opposite expression of surface biomarkers, indicative of immune cell activation or engagement. PEPSI categorizes cells into distinct subtypes: polarized, uniform, and others, based on their protein marker polarity. This classification is crucial for analyzing the functional states of immune cells within tumors.
Applied to multiple large-scale datasets encompassing over two million cells and 600 patient samples, the method has provided insightful revelations. Cells identified with polar expression exhibited characteristics relating to tumor-immune cell engagement. Incorporating these polarity-defined cell subtypes into deep learning models resulted in a notable improvement in predicting patient survival outcomes. This suggests that surface protein marker polarity is a significant factor in characterizing the functional state of immune cells.
In conclusion, PEPSI stands out as an advancement in spatial proteomics, providing new insights into the functional conditions of immune cells in the tumor microenvironment. It employs a unique approach to phenotype immune cells based on subcellular protein expression patterns. This methodology could revolutionize researchers’ understanding of patient responses to treatments and disease prognostics. The ability of PEPSI to refine the analysis of tumor microenvironments marks a significant step forward in precision medicine. It highlights the importance of measuring and incorporating subcellular polarity information into tissue microenvironment analyses, potentially leading to a more personalized understanding of diseases and patient outcomes.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our 36k+ ML SubReddit, 41k+ Facebook Community, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our Telegram Channel
![]()
Sana Hassan, a consulting intern at Marktechpost and dual-degree student at IIT Madras, is passionate about applying technology and AI to address real-world challenges. With a keen interest in solving practical problems, he brings a fresh perspective to the intersection of AI and real-life solutions.